TERÅ beads is an end-to-end DEL*-based platform to boost the performance of oligonucleotide therapeutics by deep-learning the effects of chemical modifications
It all starts with a bead
The first step of the TERÅ beads (topologically encoded, RNA-active) platform is to create a vast number of therapeutic RNA molecules with unique patterns of chemical modifications.
During the synthesis of the therapeutic RNA molecules, we also create DNA barcodes on each bead that record the pattern of these RNA modifications, making them accessible to high throughput sequencing.
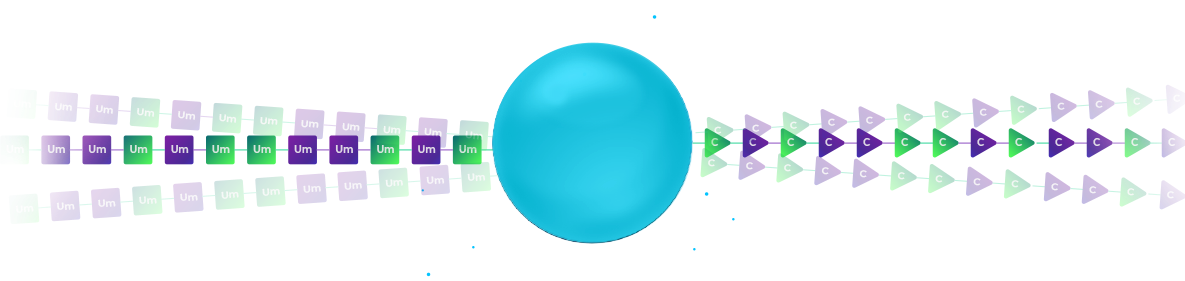
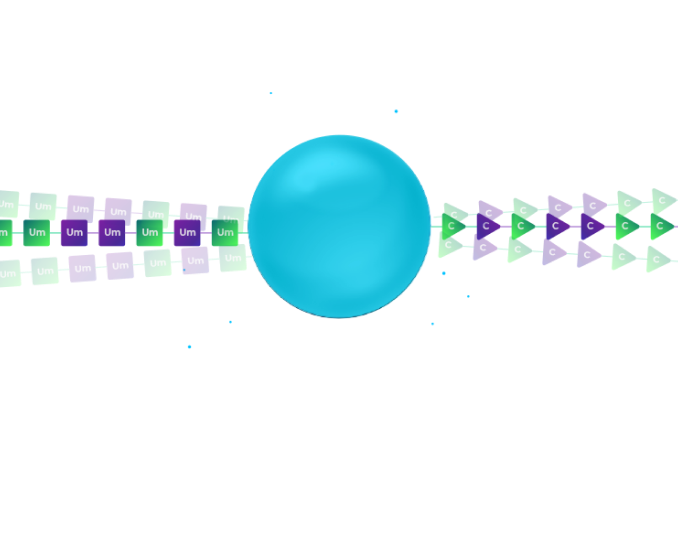
To synthesize these molecules in a cost-effective manner, we subject millions of beads to a split-pool process.






The beads are split at random into different synthesis columns.








Our patent-pending orthogonal chemistry coupling simultaneously synthesizes the modified RNA oligo and the DNA barcode.
In the illustration, the left column uses 2′-F Uridine as the first base of the therapeutic RNA oligo and a DNA pyrimidine nucleotide to signify the addition of the 2’F-U modification. The right column uses 2′-OMe Uridine for the RNA oligo and DNA purine nucleosides to signify the 2′-OMe modification of the RNA.
The process can support a wide repertoire of chemical modifications in the nucleobase, sugar ring, and phosphate backbone.
After synthesis, the beads from all columns are pooled, creating a mixture of beads.
In the illustration, we have a mixture of two types of beads: those with 2’F-Uridine and those 2′-OMe-Uridine. Each of these beads is labelled with a DNA nucleotide according to the chemical modification.
We subject the mixture of beads to an additional split. Again, in each synthesis column, we use the orthogonal chemistry to extend the therapeutic RNA oligo by an additional modified nucleoside and to record the identity of the modification by coupling the proper nucleoside to the DNA barcode in a distinct reaction.
In the illustration, the left column couples 2′-F-Cytidine and the right column couples 2′-OMe-Cytidine. Again, a coupling of DNA pyridine nucleoside signifies the left column and the coupling of a purine DNA nucleoside signifies the right column.
After two split-pool cycles, we have four types of beads.
Each synthesis path creates a distinct pattern of chemical modification for the RNA oligonucleotides on the bead and a unique sequence on the DNA barcode that records the synthesis path of the bead.


At the end of the process, each bead holds therapeutic
RNA oligos with a unique chemical pattern and DNA
barcodes that record the identity of these modifications.





The TERÅ platform allows us to refine our hypothesis space and to only advance to in vivo testing those RNA oligonucleotides that are backed by sufficient evidence from the in-cellulo assay.
RNAi triggers, ASOs, gRNA, and even short mRNA — if it can be chemically synthesized, the revolutionary TERÅ® platform can find the optimal pattern for chemical modifications. These modifications can increase the duration of effect of the therapeutic molecule, tune its bioavailability, reduce immunogenicity, and mitigate off-target effects.

